Molecula Package
The
Molecula package introduce a data structure for 3-dimensional macromolecules description, and number of functions useful to manipulate and vizualize these complex structures.
Functions in the Molecula Package
Data importation
Introduction
Molecula has its own import function to load molecular data from PDB files. This is mainly due to the introduction of a different description of molecules used within this package.
This imports a molecule from a PDB file
| Out[1]= | 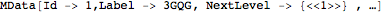 |
Molecular expressions are often very large. By default, only condensed print forms prompted. To print the complete expression use
InputForm function:
This prints the InputForm of mol (
this gives a very large output !!!)
At first load from a PDB file, an MData object is returned. The content of each level can be explored with the
MContents function:
This prompt an interface to explore the content of loaded information
This expression contain most of the information read from the file. This information is organized hierarchically following 5 levels of inclusion : Data > Models > Chains > Residues > Atoms. Various descriptors are associated to each level.
Getting pieces of molecular expressions
| MExtract | extracts sub expression according to descriptor values |
| MKeep | cleans up an expression keeping only sub expressions according to descriptor values |
Getting pieces of molecular expressions.
Two powerfull functions allows to select parts of a molecular expression. The first,
MExtract extracts from the hierarchy, elements of interest, and the second, MKeep, keeps original data structure hierarchy, keeping as sub expressions
This extracts the first model
| Out[10]= | 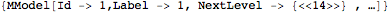 |
This extracts the first chain of the first model
| Out[18]= | 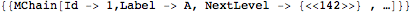 |
Curly brackets keeps track of the original level of hierarchy of extracted element, as long as these levels are specified in the selection pattern. The following evaluation keeps track only of the "residue" and "atom" level:
This extracts all residues, and then extracts all atoms with label "FE"
| Out[19]= |  |
Selection of pieces of molecular objects are made by pattern matching. Admited patterns concerns index numbers and labels of hierarchical expressions:
This extracts all residues with label "TYR" -
i.e. all cysteins - from chain
| Out[21]= |  |
Mix of label patterns and index patterns is supported:
This extracts from residues with index number in the range 1 to 10 and residues with label "CYS"
| Out[26]= |  |
Pattern of selection can be much more sofisticated, using the
Mathematica pattern functions Alternatives and Except :
This extracts atoms with labels "C", "N" or "CA", from residues with label "CYS"
| Out[23]= | 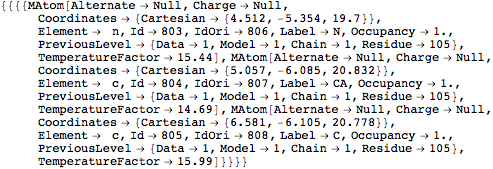 |
A special caracter is dedicated by
Mathematica to match everything:
Blank[] also denoted
_This extracts all atoms but "C", "N" or "CA", from all residues
Data structure organization
The data structure of molecule object is hierarchically structured. An analogy can be maid to a tree-graph structure. Each level correspond to a specific hierarchical expression denoted
hexpr. The different levels of organization, from global to specific -
i.e. from root to leaves- are:
- The Data level: The head of the corresponding expression is MData. When a PDB file is first imported with MImportPDB, an MData hexpr is returned. It contains various informations read from the PDB file, and a complete hierarchical description of its content. Often, when molecule structures are derived from NMR experiments, PDB files content many alternative conformations of a single molecules. Each conformation is considered as a model in Molecula.
- The Model level: The head of the corresponding expression is MModel. Biological macromolecule definition encompass the chemical definition. According to so called quaternary structure, macromolecules can be made of one or more chains. These chains are grouped into a model.
- The Chain level: The head of the corresponding expression is MChain. All atoms contained in a chain are directly or indirectly covalently bounded. A chain is equivalent to a molecule according to the chemical definition. A chain is build from residues.
- The Residue level: The head of the corresponding expression is MResidue. A convenient way used to describe polymeric macromolecules is to introduce the residue description. A residue is defined as a monomeric unit. In proteins it corresponds to amino acids, and in DNA or RNA to nucleic acids.
- The Atom level: The head of the corresponding expression is MAtom. Atoms are the leaves of the hierarchical data structure. At first, the description of atoms is limited to data stored in PDB files which describe atom type, position in a cartesian reference frame and optionaly few characteristics such as occupancy or temperature factor.
| MData | head of the container of molecular data |
| MModel | head of a model hierarchical expression |
| MChain | head of a chain hierarchical expression |
| MResidue | head of a residue hierarchical expression |
| MAtom | head of a atom expression |
Heads of Molecula hierarchical expressions.
All
hexpr are identically structured. They are made of a head collecting a collection of descriptors defined as rules:
Head[ descr1 → val1 , descr2 → val2 , ... ]There are three generic descriptors:
"NextLevel": gives the list of sub hierarchical expressions contained by
hexpr,
"Label": gives the name of the
hexpr and
"Id": gives the index number of the
hexpr.
This structure is similar from many points to an object-oriented structure. Each level is characterized by a set of descriptors and one of them gives the list of the successors.
Getting elements
Exploration, edition, modification of the data structure are done by a small set of functions. This is possible because every structration level in organized identically uppon the head/descriptor list paradigm of
hexprs.
Low level data structure manipulation functions.
This lists the descriptors defined at the first level for mol object
| Out[6]= | 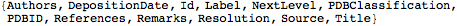 |
This lists the descriptors defined at the first level for mol object
| Out[6]= | 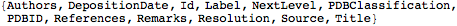 |



